The OpenADMET - ExpansionRx Blind Challenge has Come to an End
Maria Castellanos Hugo MacDermott-Opeskin
Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties can make or break the preclinical and clinical development of small molecules. At OpenADMET we aim to address the unpredictable nature of these properties through open science, generating high-quality experimental data and building robust predictive models of ADMET properties.
A key component of these efforts is running community blind challenges, designed to benchmark the state-of-the-art in ADMET models from a diverse array of participants on novel datasets. With these goals in mind, on October 27, 2025 we launched the ExpansionRx-OpenADMET blind challenge, in partnership with Expansion Therapeutics and Hugging Face.
The challenge reflected real-world data complexity
For this challenge, participants were tasked with predicting nine crucial endpoints from a real-world drug discovery campaign prosecuted by ExpansionRx targeting RNA-mediated diseases, including Myotonic Dystrophy, Amyotrophic Lateral Sclerosis (ALS), and Dementia. Comprising over 7000 molecules, the Expansion dataset is one of the largest lead-optimization-like datasets available in the public domain (more on this later) and is a highly realistic benchmark for use both in this challenge and in the future.
When designing a challenge to test the utility of machine learning models in drug discovery, it is crucial to frame it in a way that mirrors the real challenges faced by medicinal chemistry and data science teams. To achieve this, we used a time-split strategy. Participants were tasked with predicting ADMET properties of late-stage molecules using earlier-stage data from the same campaigns.
During the challenge, hosted in a Hugging Face space, participants could access a fully disclosed training set, as well as the molecules in the test set (with endpoint data blinded). After training, predictions on the full test set were uploaded to the Hugging Face platform and evaluated against a subset (half) of the test set (validation set), while the other half remained hidden. Submissions were ranked based on the macro-averaged relative absolute error (MA-RAE) with respect to this validation subset, providing a reflection of performance against the blinded test set while also discouraging participants from overfitting their models on the live leaderboard. Participants were evaluated on the full test set once at the intermediate leaderboard, and again at the close of the competition (this blog post!). We believe this represents a balance between community engagement through the leaderboard, while minimizing potential overfitting using its evaluation metrics.
Providing a new high-quality dataset for the community: Full data release
Today, we aren't just revealing the winners of the challenge: We are officially releasing the full dataset provided by ExpansionRx 🎉. You can find it on Hugging Face. We hope that this dataset will help the drug discovery community develop new predictive models and improve existing ADMET-prediction methods. In the words of Jon Ainsley, from the ExpansionRx team:
"When we launched this challenge, we asked the scientific community to put our data to work - and honestly, they delivered beyond anything I imagined. Over 370 participants brought creativity, rigor, and genuine collaboration to a problem that matters deeply, not only to Expansion, but the wider drug discovery community. This is what's possible when real project datasets meet open science. It's been remarkable to see the community's ingenuity on full display, approaches I hadn't considered, new methods shared openly, and the state of the art brought into the open where everyone can learn from it.
Now, with the full dataset released, we pass the baton to the broader community. Build on it, benchmark against it, prove us wrong about what's predictable. Every improvement gets us a step closer to a future where ADME becomes straightforward.
To others with data to share: consider publishing what you can and give the community better problems to solve. The more real-world data we collectively put out there, the faster we make drug discovery simpler, and the sooner patients benefit."
Extraordinary community engagement
The response to the challenge surpassed our wildest expectations. By the closing date of January 19, 2026, the challenge had seen:
- 370+ participants across industry and academia, with different backgrounds and levels of experience.
- More than 4000 total submissions.
- Lively collaboration in our Discord server, which became a hub for troubleshooting, method sharing, and open-source contributions from participants.
- Training data downloaded over 1,000 times.
This exercise was a true reflection of what the drug discovery community can achieve by sharing their methods through open science! We want to thank every participant for their efforts throughout the challenge. It’s been super inspiring to see all the modeling approaches and the community spirit fostered.
Over the course of two and a half months, challenge engagement increased steadily. The Empirical Cumulative Distribution Function (ECDF) plot below shows the cumulative number of new participants or teams (identified by their unique Hugging Face username) joining each day.
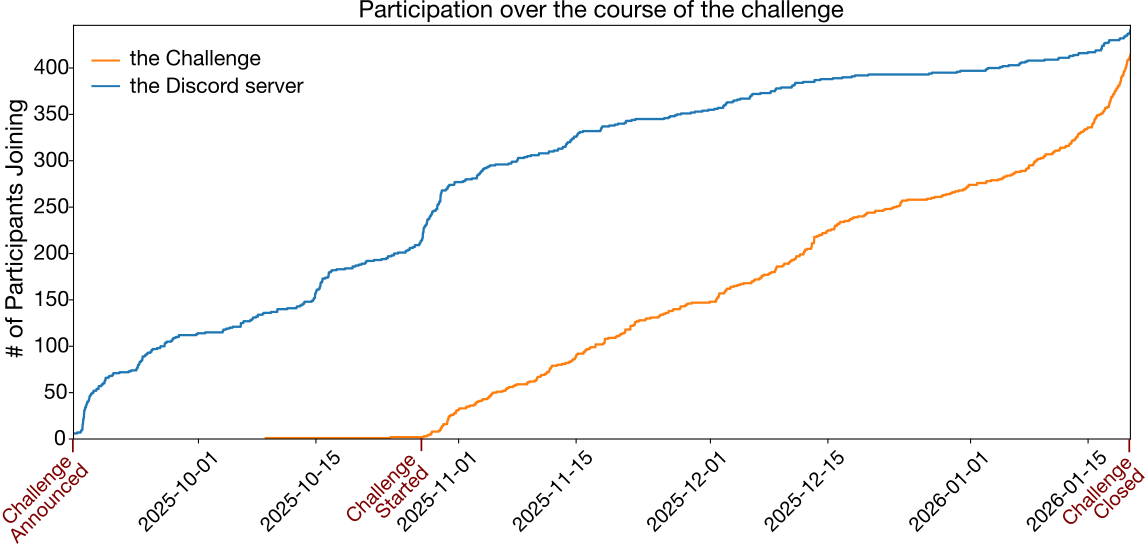
Additionally, our Discord channel proved to be a great platform for participants to engage in fruitful conversations and collaborate to build better models. A total of 446 participants joined the Discord server, starting from the day the challenge was announced.
Our evaluation strategy aimed to capture the error across all endpoints
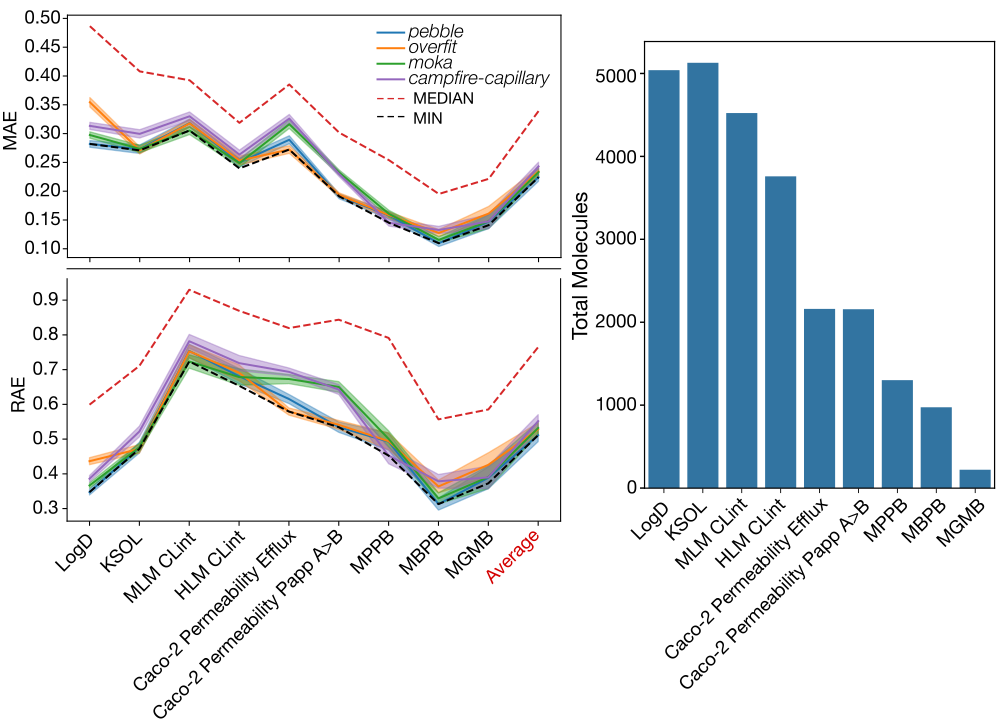
Participants were evaluated and ranked according to the MA-RAE, which normalizes the MAE to the dynamic range of the test data and gives equal weight to errors between endpoints. This is especially important when there is a data imbalance between the different endpoints.
An example of the MAE and RAE distributions across the nine endpoints is shown below for the top 4 participants on the final leaderboard. When compared to the minimum distribution for each endpoint, we see that using the average of the RAE as a leaderboard evaluation strategy managed to capture the best performers across all the endpoints, instead of biasing toward the endpoints with a larger number of molecules, such as LogD and KSOL.
Next steps
We hoped you enjoyed participating in this challenge as much as we enjoyed hosting it! Here’s what to look out for:
-
A second blog post will be posted next week, diving deeper into the trends we observed on the models and techniques used by the top participants. Please stay tuned!
-
We will host a series of webinars, during which the top participants may present their work and results if they wish. These webinars will be recorded and made available asynchronously on YouTube.
-
We plan to release a summary preprint detailing the challenge results, and all challenge participants on the final leaderboard are invited to be co-authors.
OpenADMET is committed to advancing ADMET predictive modeling and will continue to host blind challenges quarterly. Therefore we prepared this survey: Please fill it out with your contact information and any and all feedback on this past challenge. This information will be used to send invitations to the upcoming webinar series. We will be announcing our next blind challenge very shortly!
Please also indicate whether you want to be included as a co-author, and fill out your name and affiliation, as you want it to appear in the paper. Importantly, even if you had already provided your contact information with your submission, please indicate again in the survey so we have a centralized database.
The Final Leaderboard
Now, the wait is over! The final leaderboard, which shows model performance against the entire blinded test set, is shown below. Note that the live leaderboard in our Hugging Face space is evaluated only on half of the full test set (the validation set) and therefore the performance shown here is different from the live leaderboard. For easier comparison, we have included both the final Rank (evaluated on the full test set) and the initial rank (validation set). Additionally, the final rank is color-coded for clarity: green indicates the rank was maintained or improved compared to the initial standings, while red indicates the rank was lower. Note that some participants ranked higher due to those that dropped out from the competition due to not fulfilling eligibility criteria (e.g model reports), however we deemed it important to provide a link to participants' original position on the live leaderboard.
To assess whether the difference between submissions is statistically significant, we have included a Compact Letter Display (CLD) in the table below. The CLD summarizes the results of the Tukey HSD (Honestly Significant Difference) test, which compares every possible pair of group means from bootstrapped samples.
The Tukey HSD test is used to identify precisely which groups differ from one another after an ANOVA finds a general difference. The CLD translates these complex pairwise findings into a simple code:
Same Letter - Groups that share a letter (e.g., "a" and "ab") are not statistically different according to the Tukey test.
Different Letters - Groups that share no common letters (e.g., "a" vs. "b") are significantly different.
Note that the bootstrapping process used to generate performance distributions here likely underestimates model variance that would be obtained via a more rigorous process such as cross validation, however these techniques are considered beyond the scope of this challenge for technical reasons.
Important Notes
-
Invalid Hugging Face usernames:
Submissions made using invalid Hugging Face usernames (a requirement stated from the start and reiterated in the “Submit” tab) have been removed. -
Required model reports:
All submissions to the final leaderboard required a valid link to a written report or GitHub repository providing a general description of the model used. Because it was announced in advance and participants were reminded multiple times, submissions that did not include this report are excluded from this final leaderboard. This transparency is an essential component of the challenge, as it helped us, the organizers, and the community understand which models and strategies lead to better predictive performance and will help advance ADMET models in the future.
Congratulations to all participants on your efforts and we look forward to seeing you at the upcoming seminar(s) and in our next blind challenges!
Questions or Ideas?
We’d love to hear from you! Whether you want to learn more, have ideas for future challenges, or wish to contribute data to our efforts.
Join the OpenADMET Discord or contact us at openadmet@omsf.io.
Let’s work together to transform ADMET modeling and accelerate drug discovery!
Acknowledgements
We gratefully acknowledge Jon Ainsley, Andrew Good, Elyse Bourque, Lakshminarayana Vogeti, Renato Skerlj, Tiansheng Wang, and Mark Ledeboer for generously providing the Expansion Therapeutics dataset used in this challenge as an in-kind contribution.